All published articles of this journal are available on ScienceDirect.
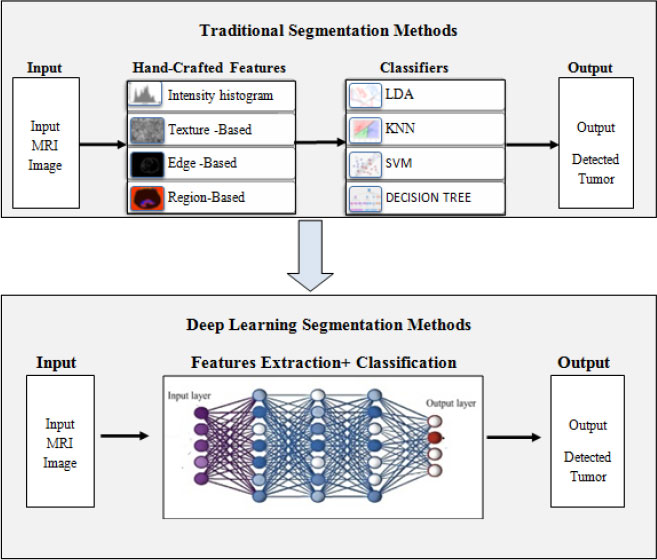
Trends in DNN Model based Classification and Segmentation of Brain Tumor Detection
Abstract
Background:
Due to the complexities of scrutinizing and diagnosing brain tumors from MR images, brain tumor analysis has become one of the most indispensable concerns. Characterizing a brain tumor before any treatment, such as radiotherapy, requires decisive treatment planning and accurate implementation. As a result, early detection of brain tumors is imperative for better clinical outcomes and subsequent patient survival. Brain tumor segmentation is a crucial task in medical image analysis. Because of tumor heterogeneity and varied intensity patterns, manual segmentation takes a long time, limiting the use of accurate quantitative interventions in clinical practice. Automated computer-based brain tumor image processing has become more valuable with technological advancements. With various imaging and statistical analysis tools, deep learning algorithms offer a viable option to enable health care practitioners to rule out the disease and estimate the population growth.
Methods:
This article presents a comprehensive evaluation of conventional machine learning models and evolving deep learning techniques for brain tumor segmentation and classification.
Conclusion:
In this manuscript, a hierarchical review has been presented for brain tumor segmentation and detection. It is found that the segmentation methods hold a wide margin of improvement in the context of the implementation of adaptive thresholding and segmentation methods, the feature training and mapping require redundancy correction, the input data training needs to be more exhaustive, and the detection algorithms are required to be robust in terms of handling online input data analysis/tumor detection and cancers.
1. INTRODUCTION
The brain tumor is a serious health-related issue that is considered a type of cancer. A higher chance of survival can be achieved with a more accurate and earlier brain tumor detection. Furthermore, it is difficult to accurately identify distinct types of tumors. The present study presents strategies for brain tumor segmentation and detection briefly and comprehensively. The motivation for the presented work includes:
- Rapid diagnosis of bugs
- High accuracy of results
- Short diagnostic time
- Assisting medical specialists in identifying and curing disease in its early stages
- Save the patient’s life and time
1.1. General Aspects of Brain Lesions and their Imaging Techniques
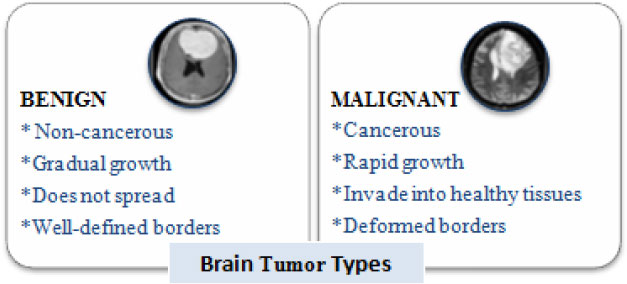
Brain tumors perpetuate because of the unusual advancement of cells that multiply uncontrollably [1]. Tumors can begin from brain cells around the membrane of the brain (meninges), organs, or nerves. The primary subtypes of brain tumors are benign and malignant tumors (Fig. 1). Benign tumors develop slowly and are non-cancerous and less invasive. On the contrary, a malignant tumor is a harmful tumor, expanding rapidly with unknown borders that invade other strong body cells. It is recognized as a primary malignant tumor if found in the brain. It is a secondary malignant tumor [2] if it originates somewhere in the body and extends to the brain. Computed Tomography, Single-Photon Emission Computed Tomography, Positron Emission Tomography (PET), Magnetic Resonance Spectroscopy, and Magnetic Resonance Imaging (MRI) are all clinical imaging modalities that are used to impart critical information regarding the form, scale, area, and metabolism of brain tumors. These modalities are used to provide the most up-to-date information on brain tumors. Due to its remarkable delicate tissue distinction and extensive accessibility, magnetic resonance scanning is the dominant methodology [3].
MRI is a non-invasive diagnostics mechanism that involves radio recurrence signs to charge tissue of interest and establish an image impacted by the advancement in recent technology [4]. Excitation and reiteration rates are adjusted upon image procurement to create pictures of varied MRI successions. These numerous MRI methods generate tissue differentiation variants pictures, imparting valuable insights and facilitating the conclusion and division of tumors and their sub-regions [5]. Furthermore, MR images provide critical information on various tissue dimensions such as proton density(PD), spin-lattice (T1), and spin (T2) relaxation times, flow rate, and chemical shift), allowing for an additional pertinent depiction of brain tissue. T2 weighted (T2) pictures are often used to give a basic assessment, distinguish malignancies from non-tumor tissues, and recognize tumor subtypes. The difference material in T1 weighted pictures (T1) aids in the delineation of tumor edges from adjacent healthy tissues [6]. Fluid Attenuation Inversion Recovery (FLAIR) is used to accomplish a T2-graded scanning in axial projection to show non-enhanced tumors [7]. MRI has a substantial advantage in brain tumor research due to these distinguishing characteristics.
Table 1 enlists some terms used in MRI. The diagnosis of a brain tumor is determined by the patient's age, the type of tumor, and the location of the tumor. Tumors can emerge and spread to the surrounding healthy tissue, making diagnosis and treatment challenging [8]. As a result, brain tumors must be concisely delineated from encompassing regions to detect tumors at a preliminary phase and increase patients' survival chances. Diagnosing, highlighting, and isolating tumor tissues such as dynamic cells, necrotic centers, and edema from benign cognitive cells like Gray Matter (GM), White Matter (WM), and CSF are all parts of brain tumor fragmentation. In a segmented image, a bright signal indicates an active field, a dark signal indicates a necrotic heart, and a medium-level signal indicates edema.
1.2. MR Imaging and Segmentation
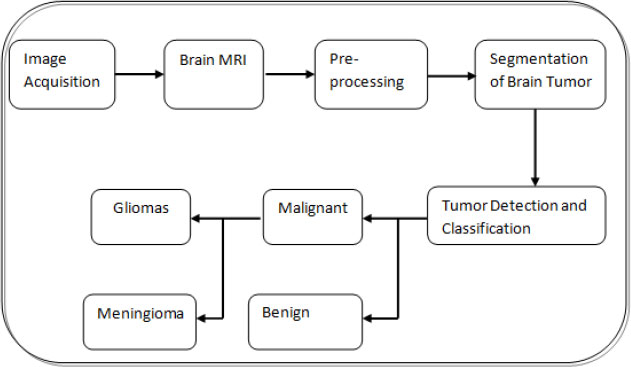
In the current clinical daily schedule, this assignment includes labeling and manually segmenting the large number of MRI scans that are multivariate. However, because manual segmentation is time-consuming, the development of autonomous fragmentation algorithms to provide proficiency and target segmentation has recently become the highest priority [9]. In Fig. (2), a block diagram of the steps involved in brain tumor diagnosis is presented. The discriminative stratification procedure involves pre-processing, extrication of features, categorization, and post-processing. Pre-processing includes noise reduction, skull-stripping, and strength bias correction [10]. Following pre-processing, image analysis procedures are used to extricate traits that closely portray distinct tissue categories. Discrete Wavelet Transforms (DWT), first-order empirical aspects, intensity variations, and edge-based traits are several instances of attributes like intensity, texture, and edge-related attributes.
| Term | Description |
|---|---|
| T1 | It is the time constant which determines the rate at which excited protons return to equilibrium. It is a measure of the time taken for spinning protons to realign with the external magnetic field. |
| T2 | It is the time constant which determines the rate at which excited protons reach equilibrium or go out of phase with each other. It is a measure of the time taken for spinning protons to lose phase coherence among the nuclei spinning perpendicular to the main field. |
| TR | It is the repetition time i.e the time between two excitations. |
| TE | It is the echo time i.e the time interval in which signals are measured after Radio frequency (RF) excitation. |
| T1-weighted image | Shorter TE and TR times leads to formation of T-1 weighted image. The contrast and brightness of the image are predominately determined by T1 properties of tissue. |
| T2-weighted image | Longer TE and TR times leads to T2-weighted images. In these images, the contrast and brightness are predominately determined by the T2 properties of tissue. |
| Flair image | It depicts the regions of tissue T2 prolongation as bright while darkening cerebrospinal fluid (CSF) signal, thus evidently highlighting the lesions in proximity to CSF. |
A wide range of classifiers, such as SVM, Neural Networks (NN), KNN, SOM, and RF, are deployed using such features. Machine learning (particularly deep learning) advancements have made it possible to locate, categorize, and assess anomalies in medical data. The use of hierarchical vectors acquired only from data, rather than manually constructed features based on specific domain knowledge, is the basis of these advancements. The use of neural networks (NNs) helps in solving complex problems, such as control problems of grid-connected solar PV systems [11, 12] and anomaly identification in wireless networks using ai (artificial intelligence) technology. Due to its extensive usage in image cryptography and secure communication, the synchronization of NNs has recently emerged as a remarkable issue. Synchronization is the technique of commanding a slave machine to track a master machine using a controller [13].
A critical stage in radiotherapy treatment is precise volume demarcation. A 3-dimensional volume of the part of the body to which a treatment dose of radiotherapy can be delivered is created using a series of cross-sectional photographs on a computer to determine the location known to have cancer and the areas at risk of tumor spread. If the area at risk is incorrectly defined, cancerous areas may go untreated, lowering the possibilities of cure. However, treating a big volume of tissue increases the amount of normal tissue treated, increasing the chance of side effects. As a result, proper identification and segmentation of tumor targets are required to apply precise radiation to the tumor while protecting the nearby tissues. In comparison to older conventional approaches, deep learning techniques have raised to the forefront approaches for brain tumor diagnosis because traditional algorithms rely heavily on manual extraction of features; in contrast, feature extraction in deep learning is automatic, where each layer of the neural network utilizes traits of the preceding layer to develop higher-level features. We hope that the results of this study will be valuable to anyone working on brain tumor segmentation and classification using deep learning. This survey contributes to the following:
(1) Brain MRI features and algorithms are reviewed and described for their qualities, benefits, and drawbacks to better understand how these tools are employed. The presented study uses threshold methods, graph-based techniques, and deep learning approaches, all the way up to deformable methods to classify segmentation algorithms.
(2) There is also an in-depth and critical examination of the most current brain image classification approaches that use DL methodologies to give a comparison study showing the precision level attained via various techniques.
The remaining paper is organized as mentioned. Section II represents a detailed discussion of various steps taken in brain tumor diagnosis, followed by a description of the traditional segmentation approaches and their related work in section III. Section IV analyzes recent trends in brain image segmentation and classification utilizing DL approaches, accompanied by a summary in section V.
2. STEPS INVOLVED IN COMPUTER-AIDED DIAGNOSIS OF BRAIN TUMORS
2.1. Pre-Processing
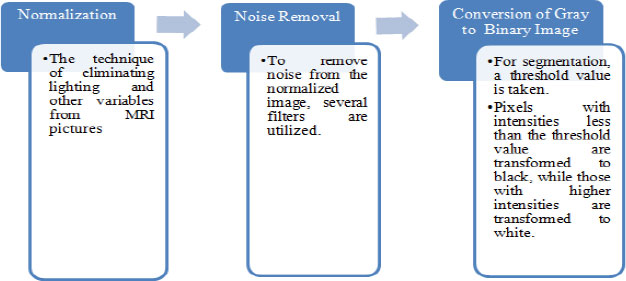
Preprocessing of an image a picture is done to lessen the noise and to improve the brain MR image for additional processing, so that the tumor can be may be identified with more certainty and ease. In this initial stage, there are three steps to be followed (Fig. 3) as referenced beneath:
A wide assortment of pre-processing procedures, like linear, non-linear fixed, versatile, pixel-based or multi-scale, are pertinent for various conditions. Exact diagnosis becomes complicated at relatively high noise levels in cases where the demarcation between normal and malignant tissue is limited [14]. A clinically trained expert will be greatly benefitted greatly with a slight increase in imaging graphics fidelity. Henceforth for resulting automated investigation, enhancing methods are amongst the necessary phases in the pre-processing procedure. Techniques for augmenting are employed for two purposes. First and foremost, to create better images that can be viewed by humans, this includes noise reduction, brightness enrichment, and refining information in a picture. Secondly, to create images that can be used in later data processing procedures comprising edge recognition and object segmentation methods.
The bias field is a major issue that arises during the segmentation of MR images. It is called intensity non-uniformity because it is caused by inconsistencies in the procurement phases or radiofrequency loop abnormalities [15]. Bias field correction aims to compute and remove the bias field from the picture [16]. Wang et al. explain the bias correction criteria in the pre-processing stage of an MR image [6]. As a result, edges and features are not sufficiently recuperated, particularly at high noise levels. According to image processing experts, median filtering is a better alternative for reducing noise than linear filtering when edges are included. Fact that the data set could come from a variety of MRI scanners presents another hurdle for the algorithms. Since MR image intensities vary between MRI scanners, this set contains MR images of various intensities. Other concerns include different types of noise produced by MRI scanners, inter-slice strength variations, tumor-related issues while aligning and recording images, and so on. To remove these issues, a variety of pre-processing steps are used. Focusing on the pre-processing step of MR images before sending them to the classifier is crucial; otherwise, a fault in the pre-processing step could cause the entire system to fail [17]. In their article, Tanzila et al. propose two brain extraction algorithms for T2-weighted MRIs: 2D Brain Extraction Algorithm (BEA) and 3D-BEA [18]. The purpose of brain MR image extraction using T2-weighted statistics is to reduce the size of the MRI file and, as a result, the network application delivery latency. To extract brain tissue from undesirable characteristics, BSE, BET, Hybrid Watershed methodology, McStrip, and other models were proposed by Ortiz et al. [19]. In terms of robustness and accuracy, many semi-automated and automatic brain extraction approaches are not suitable due to the occurrence of exclusion and inclusion errors [20].
2.2. Segmentation
The next move is to segment the brain tumor MR image after enhancing the brain MR image. Segmentation is used to distinguish the foreground and background of an image. Segmenting an image often reduces the amount of time it takes to process subsequent operations on the image. Although human eyes can quickly identify and isolate objects of interest from background tissues, algorithm formulation is a challenging task. Since the subsequent steps depend on the segmented area, segmentation directs the outcome of the entire study. Segmentation algorithms use region expansion, deformable models, histogram equalization, and image identification strategies like fuzzy clustering and neural networks to modulate the strength or texture of pictures. Region-based and edge fragmentation, dynamic and global thresholding, gradient drivers, watershed fragmentation, hybrid segmentation, and volumetric fragmentation, supervised and unsupervised segmentation are all readily accessible. Segmentation is achieved by identifying all voxels or pixels that belong to the object or those that form boundaries. The former employs pixel intensity, whereas the latter employs picture gradients with high values at the edges. Because it requires pixel classification, segmentation is sometimes viewed as a pattern recognition task.
2.3. Manual Segmentation Methods
To do manual segmentation, the radiologist must employ the multi-modality knowledge offered by MRI images, as well as physiological and behavioral abilities obtained via training and experience. The radiologist examines multiple images in segments one by one, attempting to diagnose the tumor and precisely sketching a schematic of the tumor locations. Manual segmentation takes time and is heavily dependent on the radiologist, with considerable intra- and inter-rater variability [19]. Manual categorization, on the other hand, is routinely used to assess the results of semi-automated and fully automated methods.
2.4. Semi-Automatic Segmentation Methods
For three key reasons, semi-automatic methods necessitate human input: initialization, intervention or input response, and evaluation [16]. The automatic algorithm is usually initialized by identifying a region of interest (ROI) containing the estimated tumor region for processing. Adjusting the parameters of pre-processing procedures necessitates intervention. By obtaining input and making adjustments in response, an automated algorithm can be directed toward the intended outcome. Finally, the user can assess the results and alter or repeat the procedure based on the requirements. In addition, if the user is unsatisfied with the results, he or she can go back over them and change or repeat the method. The “Tumor Cut” approach was proposed by Vaishnavee KB et al. [9]. To use this semi-automated segmentation method, the user must sketch the tumor's maximal diameter on the input MRI images. To generate a tumor probability map, a cellular automata (CA) based seeded tumor slicing technique is executed twice, once for user-supplied tumor seeds and once for background seeds. This method involves running the algorithm to each MRI modality independently (for example, T1, T2, T1-Gd, and FLAIR), then merging the data to get the final tumor volume.
A novel classification approach was used for a recent semi-automatic system by Anitha et al. [17]. The fragmentation problem was translated into a classification problem in this method, and a brain tumor was fragmented by training and categorizing within the same brain area. For brain tumor segmentation, machine learning classification methods usually involve a large number of brain MRI scans from different instances to train on. As a consequence, intensity bias mitigation and other noises must be dealt with. However, in this approach, the user starts the process by choosing a subset of voxels from a single case that belongs to each tissue type. The method uses intensity values and some spatial coordinates as characteristics to identify these subsets of voxels and then trains a support vector machine (SVM) to match all voxels in the same image to their associated tissue form. Aside from the fact that semi-automated brain tumor differentiation methods are faster and more accurate than manual approaches, they are nevertheless subject to inner and trans-assessor heterogeneity. As a result, the vast of existing brain tumor fragmentation study is based on completely automated approaches.
2.5. Fully Automatic Segmentation Methods
In fully autonomous brain tumor differentiation systems, no user intervention is required. To solve the categorization problem, artificial intelligence and foreknowledge are widely employed. Liberman et al. suggested a study to increase the reliability and precision of automatically assessing treatment response in recurrent glioblastoma [21]. On 59 longitudinal MR imagery from 13 subjects, a k-Nearest Neighbor (kNN) stratification system was used to determine the changes in tumor size. This procedure was then correlated to Macdonald’s parameters and manual volumetric measurements. This approach was ideal for all scans with malignant tumors with uncertain boundaries. Even though the outputs were validated using Magnetic Resonance Spectroscopy (MRS) and by a neuroradiologist, there was a strong correlation (r = 0.96) among the manual estimations of tumor volume, but only 68 percent of them matched Macdonald’s criterion.

2.6. Feature Extraction
The method of modifying or transforming an image into a set of characteristics is called feature extraction. Texture features, co-occurrence matrix, Gabor features, wavelet transform, transform dependent features, decision boundary feature extraction, minimal noise fraction transform, and non-parametric weighted feature extraction are all examples of feature extraction methods. Principal component analysis (PCA), linear discriminant analysis (LDA), and independent component analysis (ICA) are used to reduce the number of features. When feature extrication and feature reduction techniques are combined, precise systems with fewer attributes that can be extracted at a lower computational complexity are developed [22, 23]. The features employed for brain tumor segmentation are governed by the tumor type and grade, which are the two most prominent aspects. This is because different types and grades of tumors have different appearances, which may include shape, position, regularity, contrast influx, and so on. The image intensities are the most widely used features, implying that different tissues contain differing grey levels. Local image textures are another common feature since various parts of the tumor have distinct textural characteristics. The alignment-based characteristics rely on previous spatial experience. The use of such a blend of alignment-focused and textural features enhanced the efficiency tremendously. Edge-based features or strength gradients may be employed to grow a contour towards the tumor frontiers [24]. In evaluating gliomas, non-pictorial diagnostic factors such as calcification, blood supply; hemorrhage, edema, and age have become significant. MRS traits or a mixture of photometric and textural data have been employed in recent research to distinguish between different types of brain tumors. Whether using the most up-to-date classifiers or predictive analysis techniques, MRS features have been shown to improve the accuracy of brain tumor localization [25]. Few processes rely on relatively large features, making memory storage challenging [17]. The extraction of features is an essential step in the segmentation process since extracting feature sets can be difficult due to the wide variation in characteristics from one image to the next [26]. However, using all heterogeneous data results in feature vectors with a lot of dimensions, which reduces the device’s accuracy significantly. As a result, a reliable feature selection approach must be used to make accurate brain tumor descriptors of substantial strength that reduce irrelevant variables.
Even though kernel-based approaches are less susceptible to high-dimensional input spaces, further dimensionality reduction improves categorization accuracy [27]. SVM is a well-known method for dealing with small datasets and input spaces with multiple dimensions. As explained by Ortiz et al., a number of the retrieved features may worsen the classifier’s output, and all of the features may not be sufficiently discriminating on all of the pictures [26]. The method of feature extraction and deciding is critical in determining segmentation results. Principal Component Analysis (PCA), kernel PCA, and ICA support lowering of dimension [28], whereas Genetic Algorithm, Sequential Backward Selection (SBS), Sequential Forward Selection (SFS), and Particle Swarm Optimization (PSO) are popular feature selection algorithms [23]. By that the effect of the curse of dimensionality, learning models function better when features are chosen, which accelerates the learning procedure, improves generalization capabilities, and improves model interpret-ability. The feature space gets highly dimensional if this platform is skipped, resulting in poor classifier output [29].
2.7. Classification
In certain systems, the fragmentation dilemma is the translation into a stratification challenge and a brain tumor is segregated using learning and categorizing. In general, to train on huge volumes of magnetic Resonance scanning with documented underlying data from many instances, a supervised deep-learning methodology for brain tumor segmentation is necessary. To solve the segmentation problem, AI and foreknowledge are often blended. DL techniques [30] are currently used to capture better-segmented results. The accuracy of stratification, the speed of the algorithm, and the amount of computing power available are the factors to consider when designing an optimal classifier [22]. Unsupervised classification approaches such as Self-Organizing Map (SOM) and FCM are used to classify brain MRI data using supervised approaches such as ANN, SVM, and k-NN. Kharrat et al. suggested a heuristic technique for brain tumor stratification [22]. GA and support vector machines were used to stratify the data. Wavelet-based texture feature was extracted. The spatial grey level dependency approach (SGLDM) was used to extricate the traits and then the retrieved trait was fed into the SVM classifier as input. GA resolves the classification problem of feature selection. The work [22] reports an accuracy rate ranging from 94.44 percent to 98.14 percent. Fig. (4) shows a flowchart for diagnosing brain tumors using a common computer-aided diagnostics (CAD) scheme. The tumor detection stage is skipped since the computer-aided diagnosis method assumes the tumor exists in the gathered samples.
Chandra et al. developed a Particle Swarm Optimization-based clumping approach [10]. The proposed method was contrasted to SVM and AdaBoost in extracting brain tumor patterns against MR imagery. Several PSO controlling parametric quantities produce better results, as does an algorithm that determines the centroids of a cluster, connecting the brain pathways collectively. Support Vector Machine (SVM) and AdaBoost were used to compare the outcome. The investigation revealed that the proposed method's qualitative results were comparable to SVM. A reliable strategy for determining the location of a brain tumor and extracting the region of the tumor was proposed by Mehmood et al. [31]. The suggested technique used nave Bayes classification to diagnose brain tumors using MR images. Clustering via K-means and demarcation recognition approaches were used to identify the brain tumor regions. This approach yielded a diagnostic accuracy of above 99 percent. Priya et al. intended to analyze brain tumor imagery based on their grades and kinds using SVM as the stratification strategy [32]. This section focuses on tumor types such as normal, glioma, meningioma, metastasis, and four categories of astrocytomas. The SVM classifier in this work used 1st order, 2nd order, and both order features. The results demonstrate that for tumor types and grade, classification using the 2nd order trait had a precision of 85 percent and 78.26 percent, respectively. At the same period, the accuracy of the first-order features was 65.517 percent and 62.31 percent, respectively. By incorporating both, the accuracy was 84.48 percent and 68.1 percent, respectively. The findings revealed that SVM is good at classifying brain tumor types but not very well at distinguishing tumor grades. Anitha et al. described a method that uses a two-tier optimization technique for classification and an adaptive pillar K-means method for segmentation [17]. Discrete wavelet transform-based wavelets are used to extricate traits in the suggested method, which are then trained employing a self-organizing map. The K-nearest neighbor algorithm is then used to train the resultant aspects. There are two steps to the testing. In a double training phase, the two-tier classification method classifies brain cancers. The segmentation method used discriminates between normal and anomalous MRI scans. MATLAB R2013a is used to implement the system. It is demonstrated that the suggested system outperforms other standard strategies in terms of absolute performance and accuracy.
3. SEGMENTATION ALGORITHMS
To detect the tumor region, fragmentation is the procedure of breaking down an image into several segments. In this paper, different segmentation approaches based on grey level, model-based, feature texture-based, and hybrid segmentation (Fig. 5) are discussed, and these segmentation methods are summarized toward the end of this section.
3.1. Based on Gray Level
Region-based, edge-based, and thresholding algorithms fall into this group of segmentation algorithms. These are basic methods that are not always used for segmentation on their own [33]. To isolate a related area, region-based segmentation looks towards homogeneity and clusters the pixels. It involves a seeding point as an input and stretches across the volume by contrasting neighboring pixels. It is categorized into regions combining and splitting (Watershed and seed region). Gray histogram or gradient-based techniques are being used in edge-based segmentation methods. Only the outer boundary values are represented by these methods. As a result, edge-based techniques are rarely adequate for segmenting tumor regions on their own. To remove the tumor area from brain MRI, researchers combined an edge-based approach with the Watershed algorithm [34]. Even though this formulation produced promising performance, the technique only succeeds well with high contrast images and, because of the limited gradient magnitude, lacks consistency with low contrast images. Thresholding-based segmentation creates binary ima-ges from grayscale images based on a threshold value, which is a more straightforward and multipurpose solution. The pixel range extracted from the input image is used in this segmentation technique. Pixel range refers to the pixel intensities that aid in distinguishing abnormal from normal brain regions [35]. This isolation assists in the extraction of the tumor area, which benefits tumor detection research. Local and global thresholding are the two types of thresholding that are described by intensity values. Histograms are used to describe pixel intensity at all stages of intensity in local thresholding. In the global thresholding technique, the threshold value is set in beforehand, with pixels serving as cornerstones. Banerjee et al. defined a new multi-level thresholding-based region of interest segmentation method [20]. Glioblastoma multiforme tumors are detected in two stages using MRI. To begin, the segmentation is performed by searching for multiple intervals using discrete curve evolution (DCE). Then, across the signi-ficant points, a threshold value is chosen [36, 37]. Second, the fragmented imagery is pre-processed to remove each ROI centered on a manually selected predetermined set. They assert that their method is reliable, efficient, and minimizes user interaction, which is not entirely true because the seed is chosen manually, and human interaction is still needed. Although grey level strategies can extract the tumor's shape [18, 38], they are less reliable than other methods because they depend solely on the gray values of each pixel [39]. Gray level frameworks usually require human intervention, making them untenable for medical applications unless they are incorporated with other sophisticated algorithms [40, 41].
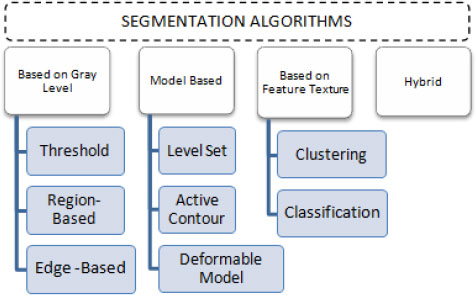
3.2. Model-Based
Model-based techniques detect the existence of contours or patterns in 2D/3D and fragment the imagery based on how well the model suits the imagery. This segmentation approach is well suited for medical imaging with repeating tissues with common traits [33], but it can cause a problem when models are rotated. A predictive approach, a probabilistic approach, a committed approach, or a blend of techniques can be used in the model. Variance, standard deviation, averaging, and other properties are used in the statistical method. Density function, data variance, normal distribution, and other techniques are often used in the probabilistic method. Deformable model [42, 43], active contour [44-46], and level-set [30] are some of the design strategies. The key disadvantage of the model-based approach is that it necessitates manual intervention, so manual seed selection or validation of the results is required in all cases [47]. Gao et al. devised an active contours-based semi-auto-matic algorithm [47]. In the image domain, the user first segments and draws the seed. After that, look into the features of these areas. Online features learning, which is local rigorous analytics performed for each voxel concerning strength, is used to extract object features. The contour (3D surface) then develops independence by using cut-off probability density and conformal metric to enclose more related tissue. Model or surface leakage is another drawback to model-based segmentation. For example, if two regions with identical intensities overlap, the segmentation result would be incorrect. The way to solve this issue is to enable manual multi-object segmentation in both regions to discriminate between them [47]. In particular, in model-based segmentation, changing the region boundary is an issue. These boundary changes could have been ensured by a partial differential equation (PDE) scheme of active contours that employs level set methods to describe area boundaries as level sets [44]. As a result, users can secure the boundary without actually understanding mathematical information or using mouse movement. But on the other hand, in image segmentation techniques that could improve the segmentation outcome, extracting relevant features of each pixel/voxel is critical. In different brain MRI modalities, for example, content-based strength and texture pattern details would be used to construct a visible curve approaching a tumor margin that is mostly homogeneous [45]. Gray Level co-occurrence Matrix (GLCM) can also be used to figure out the texture of tumor segmentation tissue. This approach produces reliable results since it couples model-based approaches with many other sophisticated algorithms to improve automaticity [42].
3.3. Based on Textural Features
To make the segmentation process more stable, precise, and simple, this approach groups pixels/voxels into regions with similar texture or intensity properties. This approach is divided into two categories: grouping (e.g. ANN, SVM, and KNN) and clustering (e.g. FCM, SOM, and K-mean). The classification process (called supervised) involves techniques to partition the data based on known labels that get trained before the segmentation by using other methods or manual methods. The simplest form of a classifier is the nearest neigh-bor classifier. In their piecewise-constant model, Harini and Chandrasekar used the nearest neighbor classifier to divide the image into regions [48]. They used kernel graph cut optimizations, which employ a Gaussian generalization model, to locate region boundaries in single and multi-dimensions without intervening humans. The nearest-neighbor classifier uses training data to position each pixel in a similar format based on its strength. To compare pixel intensities, they use a mixture model. The intensity distributions between the two regions are evaluated using the Bhattacharyya distance to determine the amount of correlation between them. Their scheme is effective, as they assert, but it is heavily dependent on the number of training photos, which is time-consuming.
Zikic et al. provide another illustration of classification [49]. They presented a framework for automatically segmenting high-grade gliomas that combines decision forest classification with context-aware traits from multimodal MRI. They used a Gaussian mixture model as an additional generative model to differentiate tumor sub-compartments from multi-modal images. Since multi-label classifiers simultaneously identify tissues, they assert that their method improves tissue component classification accuracy. Furthermore, using context-aware features removes the use of a pre and post-processing stage that uses spatial regularization to impose smoothness constraints. Even though the classification methods lead to success, training the images takes a long time. Without any training data, the clustering method (also known as unsupervised clustering) clusters data that share similar information based on pixel/voxel characteristics. Similarity indicators such as size, connectivity, and strength are used to identify clusters. To re-group the data, entails pre and post-processing. Other algorithms can be used to isolate the ROI after grouping. Self-Organizing Maps (SOM) is a clustering method that uses neural network models to group identical data items to minimize the dimensionality of MRI data [50-52]. Some study utilizes a segmentation approach based on Growing Hierarchical Self-Organizing Map(GHSOM) and a trait detection method based on multi-objective to optimize segmentation efficiency [51]. GHSOM is a type of dynamic multilayer hierarchical network topology that can be used to retrieve significant hierarchies in data. Recent research has combined SOM with the Genetic Algorithm (GA) to segment brain MRIs [52]. The relationship between the output space and the input space is used to describe boundary clustering. Feature extrication, trait selection based on GA, and voxel aggregation leveraging SOM are the three stages of the process. The use of a combination of clustering and other algorithms improves automaticity and results. For tumor fragmentation and extrication in MR images, multi-cluster optimizations such as k-means [53-55] and Fuzzy C Means (FCM) strategies [53, 56, 57] are also used. The K-mean algorithm (also known as a hard cluster) allocates pixels/voxels only to one cluster, with the number of segments determining the output. Because of the random existence of initial values, the FCM (also known as a soft cluster) is chosen because it enables partial affiliation of pixels/voxels to different clusters.
Clustering-based segmentation fragments an image into segments based on pixels of correlated intensity values. Without assisting the training images, these pixels with the same intensity values form clusters or regions. The advantage of these approaches is that they perform the training process with the available image dataset. A useful clustering method for segmenting brain images is proposed by Maiti et al. [34]. For image testing, the performance level of the fragmentation procedure was improved. Fuzzy c-means [38] and K-means clustering [41] are commonly used clustering techniques. Clumping methods such as fuzzy c-means [38] and K-means clustering [41] are widely used. The segmentation of images is classified into k groups using K-means. The mean intensity values measured for each class are used to divide these classes. The representation of each pixel with the nearest centroid value is used to segment it [53]. The key disadvantage is that it can produce weak and erroneous performance, lowering accuracy. As a result, it is regarded as a rigid and incorrect grouping. The fuzzy c means the technique is the most popular one. This procedure presents several groups based on the pixel values. Constructing c-clusters from the image is done in the regular FCM process
The K-mean algorithm (also known as a hard cluster) allocates pixels/voxels only to one cluster, with the number of segments determining the output. Because of the randomness of seed values, the FCM (soft cluster) is chosen because it permits partial adherence of pixels/voxels to multiple clusters. Sehgal, Aastha et al. used circularity as a prerequisite for extracting a tumor after clustering has been completed [56]. In the case of a linear form, this circularity-based extraction is insufficient and will be incompetent to accurately detect the tumor.
3.4. Hybrid Segmentation
Each of the approaches in the preceding categories has its own set of limitations. Hybrid segmentation, which is a blend of two or more of the existing segmentation algorithms, is used to solve these limitations. By applying the level set algorithm for demarcation of brain tumors based on gradient and intensity, Dawngliana, Malsawm, et al. [40] addressed the problem of using threshold, which neglects the tumor's characteristic. For more accurate and robust segmentation, the level set algorithm is combined with the FCM algorithm [58]. In addition, Rajendran et al. use the FCM to generate an initial contour that is being used by the deformable model to evaluate the final contour for the precise tumor boundary [30]. The k-mean approach, on the other hand, is often employed for more reliable outcomes. Siva et al. [59] and Vishnuvarthanan et al. [1] used the k-mean and fuzzy k-mean with the SOM classifier for clustering. For better outcomes, a variety of studies employ various algorithm combinations. For example, ANN is used in conjunction with Watershed [60] or Grow Cut [29], and the level set is used in conjunction with active contour. One of the major benefits of using hybrid segmentation is that it enhances the automaticity of the process, limiting the amount of human involvement. Another benefit of hybrid segmentation approaches is that the precision of the segmented tumor is elevated. However, since each paper used different accuracy metrics, comparing the accuracy of the surveyed papers that use hybrid segmentation was difficult. For their accuracy level, some of the papers used performance metrics like the similarity coefficient and Jaccard [61]. Others use standard accuracy, which is termed as the ratio of the number of pixels partitioned by the algorithm to the overall number of pixels in the image [60]. The experimental results show that adopting hybrid segmentation yields high rates in both measurements. Hybrid SOM and FCM algorithms, for example, have a normal accuracy of 96.18 percent [1], while hybrid ANN and watershed algorithms have a normal accuracy of 98 percent [60].
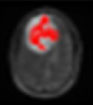
Now based on the discussed segmentation algorithms, which are used by various researchers [56, 57, 62-64] in their work for tumor diagnosis, an analysis is done in Table 2 for better visualization of the readers, where the second column depicts the data sets used followed by their choice of segmentation algorithm and then input MRI imagery taken, then segmented and detected resultant images. Also in Table 3, various pros and cons are discussed among the existing segmentation algorithms.
4. DEEP LEARNING ALGORITHMS
According to experts, deep learning is a rising subfield of ML algorithms. Instead of employing traditional attributes, DNN can acquire multilayer characteristics from the input images. Fig. (6) depicts a rough contrast between the conventional and DNN-based brain tumor fragmentation framework. Deep learning approaches are used to generate automatic features. The idea behind the concept is to run an image through a developed sequence of deep neural networks constructs before segmenting the input image using deep features. DNNs are quite efficient in automatically extricating the entire brain tumor and intra-tumor regions. In recent years, scientists have used several Deep Learning building blocks for segmenting brain tumors. Deep convolutional neural networks, RNN, long short-term memory, DNN, autoencoders (AE), and GAN are just a few examples of these types of blocks. In terms of these building blocks, the subsequent sections analyze the current literature.
4.1. Deep Neural Networks (DNNs)
DNNs are a form of neural network with multiple layers. DNN concentrates on how data is processed through a range of nonlinear capacities till it reaches the specified layer. Havaei et al. use a unique DNN model that takes into consideration both upper and lower level attributes [61]. According to scientists, the technique is faster than cutting-edge when GPUs are used.
| References | DataSet | Segmentation Algorithm | Input Image | Segmented Tumor | Detected Tumor |
|---|---|---|---|---|---|
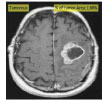
| Chithambaram et al., 2016 [56] | MICCAI 2012 | Watershed |  |
 |
 |
| Kulkarni et al., 2020 [57] | Kaggle | Threshold |  |
 |
 |
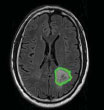
| Ji C et al., 2015 [62] | Huashan hospital, Shanghai, China | GrowCut | 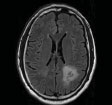 |
 |
 |
| Rajan et al., 2019 [63] | ANBU hospitals, Madurai | Hybrid (KMFCM + ACLS) | 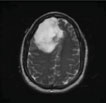 |
 |
 |
| Abdel et al., 2015 [64] | BRATS | Hybrid (KIFCM+ Threshold + ACLS) | 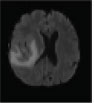 |
 |
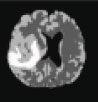 |
Table 3.
| Methods | Pros | Cons |
|---|---|---|
| Threshold-Based | No need to comprehend anything about the picture beforehand. | When images have flat or deep valleys, it becomes more complicated. |
| Region-Based | When seeds are carefully crafted, the performance of the resulting system is superior to other approaches. | Inaccurate seed selection may also result in faulty segmentation. |
| Watershed | When continuous boundaries are picked, stable and reliable results are achieved. | The problem of under segmentation or over-segmentation |
| K-means Clustering | As smaller k values are used, it works fast. | When a fixed number of clusters are taken into consideration, predicting k values becomes problematic. |
| Fuzzy C means | Better performance than K-means | Ascertaining the fuzzy membership function is a challenging task. |
| Level set | From the extracted complex shapes, pattern recognition becomes easy. | It is a time-consuming procedure because it involves manual parameter estimates. |
| Active contour | By ensuring precise simulations, accurate results can be achieved. | Noise sensitivity |
| Hybrid methods | Since it is a hybrid approach with the benefit of multiple models, more consistent results are obtained. | The complexity of computations adds to the cost. |
4.2. RNN/ LSTM
Recurrent neural networks can understand the interpretation of time sequence inputs. RNNs have a memory feature that recognizes and regurgitates information learned previously. Bi-RNNS and long-short term memory (LSTM) are examples of variations that have outperformed others in applications like video comprehension and visual question response. Most RNN-based brain tumor fragmentation uses 1D in MRI or CT volumetric statistics as the temporal dimension, and the sequential inputs of the RNN network are sections formed by either of the other 2D. RNNs are a sort of NN that operates with statistics in a sequential manner. The BraTS-17 dataset was chosen by Grivalsky et al. for HGG segmentation using the proposed RNN architecture [53]. LSTMs are a more advanced version of RNNs that are employed in sequence data design [65, 66]. Every LSTM module examines a pixel and receives data from the others. Iteratively delivers data for all distinct pixels in the graphic in this way. There are only a few studies that use LSTMs to segment brain tumors. Stollenga et al. suggest epic PyraMiD-LSTM structures for tumor segmentation that use a peculiar topology [67]. The method is easier to parallelize, needs fewer calculations overall, and works much better with GPU layouts and 3D images. On the MRBrainS13 dataset, better segmentation results were obtained. Multimodality-based segmentation is used in LSTM-MA [68]. To perform semantic segmentation, the LSTM classifier considers pixel-wise and super-pixel features. On the BrainWeb and MR BrainS datasets, the approach is evaluated.
4.3. Auto Encoders (AE)
Another DL building block is AEs. Researchers use several different AE variants to fragment brain tumors [69, 70]. In a study, 3- layers of stacked de-noising Auto Encoder were used to replicate the input dataset for fragmentation [71]. Another distinct study used a DSEN i.e, deep spatial auto encoding methodology, to segment the brain tumor. Several works focus on autoencoders [72-75]. Table 4 highlights the benefits and pitfalls of the various deep learning approaches adopted by the researchers [3, 43, 76-79] in their work.
4.4. Convolutional Neural Network (CNN)
CNN models become increasingly perplexing as structures of more than 100 layers constitute a large volume of weights and billions of correlations between neurons [80, 81]. Convolution, aggregating, activation, and forecasting are the layers that make up a typical CNN template. The literature covers a wide range of works that use CNNs to segment the brain. Mostefa Ben Naceur et al. established a three-stage pipeline to upgrade the prediction of tumoral regions in Glioblastomas (GBM) [82]. In the 1st phase, they designed deeper CNNs, and then multi-dimensional traits were extricated from significantly greater estimates of CNNs in the second phase and fed the retrieved traits of CNNs into various standard machine learning techniques such as RF, Logistic Regression, and PCA-SVM. They operated with the BRATS-2019 database. For the entire tumor, tumor center, and stimulating tumor, the average Dice value of their pipeline was 0.85, 0.76, and 0.74, respectively.
Milletari et al. suggested a unique technique for dealing with specified divisions by using the deliberation attributes of CNNs [83]. The approach is based on Hough casting, which is a strategy that considers fully programmed segmentation. To address the problem of brain tumor separation, three distinct 3D CNN models are investigated by Zhang et al. [62]. Two complete 3D CNN prototypes based on two popular 2D models for non-specific image fragmentation are proposed. A two-pathway Deep Medic variant is also being developed as a third model. In their CNN model for tumor segmentation, S Pereira et al. suggested Leaky Rectifier Linear Module [84]. A fully configured DL technique known as Input Cascade CNN [84, 85] is used to separate the tumor. Because of its two-route image preparation, an intriguing CNN design stands out from other conventional CNNs.
4.5. GAN
GAN is a CNN variant that generates high-quality data from small datasets. A generator and a discriminator are usually used to build generative adversarial networks. The first attempt is to identify the model from which the information is derived, resulting in the creation of images from noisy inputs. The later stage uses classic CNN to differentiate between actual data and data generated by the generation stage. The auto-encoder, on the other hand, helps to recreate imagery of a healthy brain from the training sample. The divergence between the stable reconstruct output and the reference value is alluded to as the segmented tumor.
| References | Method | Pros | Cons |
|---|---|---|---|
| Zhang et al. 2017 [76] | Fully convolutional neural network | When compared to CNN's initial design, it demonstrates a powerful and efficient distinguishing ability. | While detecting enhancing tumors, it makes higher false-positive predictions than intended. |
| Ibrahim et al., 2018 [77] | Fractional Wright energy function (FWF) |
More effective than the gradient-descent method in lessening the energy function. | High computational complexity. |
| Mittal et al., 2019 [3] | Stationary wavelet transform + Growing deep convolutional neural network (GCNN) | The proposed blend was proved to be more accurate than each automation approach individually. | - |
| Soltaninejad et al., 2017 [78] | Feature extraction: Intensity-based + Gabor textons + Fractal analysis Classification: Extremely randomized tree (ERT) |
Fully-automated process. | Time-consuming and also not appropriate for lesions of small magnitude. |
| Shehab et al., 2020 [43] | Deep residual learning network (ResNet) | Overcomes the gradient diffusion problem in the deep neural network through shortcut connections in the ResNet model. | An alteration in the model or a system variable is utilized to identify LGG brain tumors. |
| Khan et al.,2020 [79] | Convolutional neural network (CNN) | When compared to other pre-trained models, it consumes minimal processing power and produces significantly better precision results. | The small dataset used and proposed model can handle only binary classification problems not categorical. |
| References | Data Set | Aim | Feature Extracted/Techniques | Segmentation Algorithms | Accuracy |
|---|---|---|---|---|---|
| Shehab et al., 2020 [43] | BRATS 2015 dataset | To detect Low Grade GLIOMAS. | Intensity based features | Deep residual learning network (ResNet50) | Accuracy - 83% (Complete tumor) Accuracy - 90% (Core tumor) Accuracy - 85% (enhancing regions). |
| Rathi et al., 2012 [2] | 140 Brain MR images from IBSR | The tumor is identified as white matter, grey matter, abnormal, or healthy. | Intensity, shape, and Texture feature using LDA and PCA | SVM | The accuracy of PCA+SVM is 98.87%. |
| Mittal et al., 2019 [3] | BRAINIX Medical Images | To boost the automated system's segmentation efficiency. | Stationary Wavelet Transform (SWT) | GCNN | PSNR- 96.64% and MSE-0.001%. |
| Kasar et al., 2021 [5] | FIGSHARE Dataset | To show the effective use of semantic segmentation networks in automatic brain tumor segmentation. |
Not mentioned | UNET and SEGNET | For UNET and SEGNET, the average dice similarity coefficient is 0.76 and 0.67, respectively. |
| Vidyarthi et al., 2015 [82] | 150 Maligant brain images collected from SMS Medical College Jaipur, Rajasthan, India | Analyze the efficacy for the classes of malignant tumor types. |
Gabor wavelet and DWT | KNN, SVM and BPNN | The blend of Gabor-Wavelet + CVM + BPNN yields the best results, with a 97 percent high accuracy. |
| Sultan et al., 2019 [92] | TCIA | To delineate amongst various brain tumor classifications (meningioma, glioma, and pituitary tumor) and associated grades | Not mentioned | CNN | Accuracy of 96.13% for classifying tumor types and 98.7% for distinct grades. |
| Saba et al., 2020 [18] | MICCAI | To predict the glioma or healthy images. | Blend of shape and texture features with VGG19 | KNN, LDA, SVM, Ensemble, DT and LGR | Amongst all the BRATS datasets, optimum evaluation results came with accuracy = 0.9967 and DSC = 0.9980 in BRATS 2017. |
| Priya et al., 2016 [32] | 208 MR images for Brain tumor type and 213 for tumor grade from Harvard Medical School and Radiopedia |
To differentiate between 4 types of brain tumors—Normal, Glioma, Meningioma, Metastasis, and 4 grades of Astrocytomas. | 1st order and 2nd order statistical features using GLCM | Multi-class SVM | 68.1% -Tumor grade 84.48% -Tumor type |
| Joshi et al., 2010 [64] | TMH | To recognize tumor lesions and categorize tumor types. | Texture features using GLCM | ANN | The proposed methodology assigns a tumor grade for the Astrocytoma kind of brain cancer. |
| Ain et al., 2010 [31] | Holy Family Hospital and Abrar MRI&CT Scan center Rawalpindi | Isolation and identification of a brain tumor section. | DCT traits using K-means clustering | Naïve Bayesian classifier | 99% |
| Vaishnavee et al., 2015 [9] | IBSR from the Massachusetts General Hospital | To retrieve unusual features from medical images. | SOM clustering with Histogram Equalization |
Proximal Support Vector Machines (PSVM) |
92% |
| Khan et al., 2020 [93] | BRATS 2015 dataset | Fully automatic brain tumor segmentation. | Mean intensity, LBP and HOG | CNN | DCS - 0.81 for complete tumor DCS-0.76 for core tumor DCS-0.73 for enhancing tumor |
| Siva et al., 2020 [59] | BRATS 2015 dataset | Brain tumor classification. | ST+WPTE | Softmax regression plus DAE via JOA | 98.50% |
| Daimary et al., 2020 [51] | BraTSdataset | Automatic brain tumor segmentation. | Not mentioned | U-SegNet, Seg-UNet and Res-SegNet | U-SegNet-91.6% Seg-UNet -93.1% Res-SegNet-93.3% |
4.6. Ensemble Methods
Numerous studies have used an ensemble of multiple DL constructs to illustrate brain tumors in MR images [63, 86]. JSL (Joint Sequence Learning) proposed by Tseng et al., is a hybrid approach to tumor stratification that incorporates diverse modalities [87]. The asserted methodology integrates autoencoders, LSTMs, and CNNs. To deal with data imbalance, the two-sided learning mechanism is used. On the BRATS 2015 dataset, the model claims to have better segmentation performance. For successful tumor segmentation, Zhao et al. suggest merging CNN and RNNs [88]. Iqbal et al. derived brain tumor area using a combination of LSTM and CNN features and evaluated it using the BRATS 2015 dataset [89]. Gao et al. propose a fusion of CNN and LSTM for 4D MRI segmentation [90]. The BRIC medical and IBIS databases are the subject of the research. Ang et al. segmented different types of brain tissues using LSTM and CNN-based approach [91]. In recent days, a lot of work has been done on brain tumor MRI picture fragmentation and classification using deep techniques [64, 92, 93-104]. Still, MRI is a difficult field with plenty of room for more exploration.
The complete analysis of the various existing works on brain tumor fragmentation and stratification is summarized in Table 5.
5. DISCUSSION
Medical specialists do manual brain tumor analysis in clinics as a standard procedure. This is a difficult task due to the variety of appearances and ambiguous brain anatomy. Consequently, the process of analyzing brain images manually becomes arduous. Automated segmentation and classification, on the other hand, make the job of neurologists easier since they aid in the ultimate decision-making process. This study presents multiple MRI brain tumor segmentation methods. Despite extensive research into many methods, including edge detection, hybrid models, region growth, and classifiers, no method has been found to segment large data sets accurately and efficiently, and not all strategies apply to every type of picture. Due to a lack of gradient magnitude, edge-based methods are only useful for pictures with high intensity. The noise has a significant impact on deformable models. Complexity is a problem with model-based algorithms, such as active contour, because the seed value may be chosen incorrectly. Classification and clustering methods are initial parameter-dependent, for example, the K-mean technique produces unique results for each run. Thus, all of these limitations may be avoided by ensembling some of the approaches, emphasizing the critical role of hybrid methods in increasing utilization.
In comparison to more traditional methodologies, deep learning algorithms have become the cutting-edge tactics for brain tumor analysis in the modern era. This study represents a review of current advancements in brain picture segmentation and classification using deep learning algorithms. By assisting in the automated acquisition of features, deep learning algorithms are advantageous in brain tumor research. This significantly reduces the time required for feature engineering as compared to manual engineering. With the advent of GPUs, calculation processes have become extremely quick. Additionally, performance improves as the amount of training data grows. Apart from these advantages, there are several drawbacks to adopting DL methods in the brain tumor area. Due to the high cost of GPUs, the DL method is quite expensive. Additionally, there is no systematic literature to guide the selection of a particular deep network design for a given brain analysis application. This work will assist researchers in determining which contemporary deep learning models are being used in brain analysis, allowing for future research to be conducted using existing deep learning approaches.
CONCLUSION AND FUTURE SCOPE
Image processing is critical when it comes to examining medical images. The method of separating normal brain tissues from abnormal tumor tissues is known as brain tumor segmentation. Various recurrent and burgeoning deep neural network segmentation techniques, as well as their benefits and drawbacks, have been addressed. A critical review of state-of-the-art approaches aids scholars and physicians not only in determining numerous possibilities for investigation but also in ascertaining an accurate tumor diagnosis. In this manuscript, a hierarchical review has been presented for brain tumor fragmentation and detection. It is found that the segmentation methods hold a wide margin of improvement in the context of the implementation of adaptive thresholding and segmentation methods, the feature training and mapping require redundancy correction, the input data training needs to be more exhaustive and the detection algorithms are required to be robust in terms of handling online input data analysis/tumor detection. Also, a recent survey of the articles, demonstrates that CNN-based architectures prove to be the most preferred technique in brain-tumor-based image analysis. Many scholars upped the size of layers in the CNN network to improve accuracy because superficial layers acquire low-dimensional traits like edges and corners of an artifact while deeper layers acquire knowledge of high-dimensional traits of an image. However, applying deep learning techniques and algorithms to the study of brain tumor images poses several specific challenges. Deep learning approaches face a difficult challenge in the form of a shortfall of significant training datasets. The training of deep learning algorithms for tumor segmentation, which is mostly done in 3D networks, necessitates layer-by-layer labeling, which is a difficult and time-consuming approach. So, to recapitulate, changes in CNN architectures, as well as the inclusion of datasets from other imaging modalities, could enhance current methods, ultimately paving the way for clinically appropriate automated tumor segmentation techniques for effective treatment. In addition, the Internet of Things (IoT) has pioneered the medical field by empowering data to be gathered using a myriad of IoT devices. So, using IoT-generated images, fully automated brain tumor segmentation can be achieved, which cogently blends handcrafted features-based methodology and CNN and can substantiate to be a viable alternative.
LIST OF ABBREVIATIONS
| Pet | = Positron Emission Tomography |
| Mri | = Magnetic Resonance Imaging |
| Pd | = Proton Density |
| T1 | = Spin-Lattice |
| T2 | = Spin |
| Flair | = Fluid Attenuation Inversion Recovery |
| Iot | = Internet of Things |
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
Dr. Ayush Dogra is the Editorial Advisory Board member of The Open Neuroimaging Journal.
ACKNOWLEDGEMENTS
Declared none.


